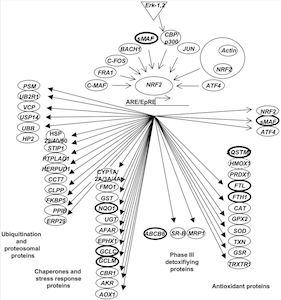
The Nrf2-Keap1 regulatory pathway regulates arsenic responsive genes
Human Health Sciences Group |

The Human Health Sciences Group conducts basic science and applied research on the mechanisms of action of the effects of chemicals and drugs in humans, how gene expression is regulated, and bone metabolism and fracture repair. We also focus on understanding the damage caused by radiation exposure, developing new technology for biosurveillance of outbreaks of infectious diseases, and accelerating the development of medical countermeasures. Our studies help us to understand how people respond to drugs and chemicals, how they vary in their response, and how to prevent deleterious effects.
DNA isolation and sample preparation for quantification of adduct levels by accelerator mass spectrometry. Dingley KH, Ubick EA, Vogel JS, Ognibene TJ, Malfatti MA, Kulp K, Haack KW. Methods Mol Biol. 2014;1105:147-57. doi: 10.1007/978-1-62703-739-6_12.
Evaluation of nanolipoprotein particles (NLPs) as an in vivo delivery platform. Fischer NO, Weilhammer DR, Dunkle A, Thomas C, Hwang M, Corzett M, Lychak C, Mayer W, Urbin S, Collette N, Chiun Chang J, Loots GG, Rasley A, Blanchette CD. PLoS One. 2014;9(3):e93342. doi: 10.1371/journal.pone.0093342.
Nanocomposite Scaffold for Chondrocyte Growth and Cartilage Tissue Engineering: Effects of Carbon Nanotube Surface Functionalization. Chahine NO, Collette NM, Thomas CB, Genetos DC, Loots GG. Tissue Eng Part A. 2014 May 20. [Epub ahead of print]
Quantifying interactions of a membrane protein embedded in a lipid nanodisc using fluorescence correlation spectroscopy. Ly S, Bourguet F, Fischer NO, Lau EY, Coleman MA, Laurence TA. Biophys J. 2014 Jan 21;106(2):L05-8. doi: 10.1016/j.bpj.2013.12.014.
The NRF2-KEAP1 pathway is an early responsive gene network in arsenic exposed lymphoblastoid cells. Córdova EJ, Martínez-Hernández A, Uribe-Figueroa L, Centeno F, Morales-Marín M, Koneru H, Coleman MA, Orozco L. PLoS One. 2014 Feb 7;9(2):e88069. doi: 10.1371/journal.pone.0088069. eCollection 2014.
Controlling the diameter, monodispersity, and solubility of ApoA1 nanolipoprotein particles using telodendrimer chemistry. He W, Luo J, Bourguet F, Xing L, Yi SK, Gao T, Blanchette C, Henderson PT, Kuhn E, Malfatti M, Murphy WJ, Cheng RH, Lam KS, Coleman MA. Protein Sci. 2013 Aug;22(8):1078-86. doi: 10.1002/pro.2292. Epub 2013 Jun 27.
D-Lactate production as a function of glucose metabolism in Saccharomyces cerevisiae. Stewart BJ, Navid A, Kulp KS, Knaack JL, Bench G. Yeast. 2013 Feb;30(2):81-91. doi: 10.1002/yea.2942. Epub 2013 Jan 30.
Directly coupled high-performance liquid chromatography-accelerator mass spectrometry measurement of chemically modified protein and peptides. Thomas AT, Stewart BJ, Ognibene TJ, Turteltaub KW, Bench G. Anal Chem. 2013, 85(7):3644-50. doi: 10.1021/ac303609n.
How genomics is changing our view of cancer. Loots GG. Brief Funct Genomics. 2013 Sep;12(5):389-90. doi: 10.1093/bfgp/elt035. No abstract available.
Interrogating transcriptional regulatory sequences in Tol2-mediated Xenopus transgenics. Loots GG, Bergmann A, Hum NR, Oldenburg CE, Wills AE, Hu N, Ovcharenko I, Harland RM. PLoS One. 2013 Jul 16;8(7):e68548. doi: 10.1371/journal.pone.0068548. Print 2013.
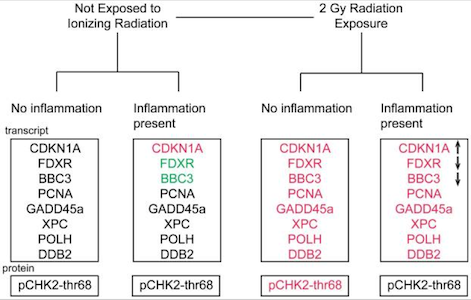
Nanosensor dosimetry of mouse blood proteins after exposure to ionizing radiation. Kim D, Marchetti F, Chen Z, Zaric S, Wilson RJ, Hall DA, Gaster RS, Lee JR, Wang J, Osterfeld SJ, Yu H, White RM, Blakely WF, Peterson LE, Bhatnagar S, Mannion B, Tseng S, Roth K, Coleman M, Snijders AM, Wyrobek AJ, Wang SX. Sci Rep. 2013;3:2234. doi: 10.1038/srep02234.
Prostate cancer invasion and metastasis: insights from mining genomic data. Hudson BD, Kulp KS, Loots GG. Brief Funct Genomics. 2013 Sep;12(5):397-410. doi: 10.1093/bfgp/elt021. Epub 2013 Jul 22. Review.
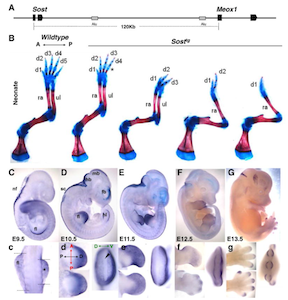
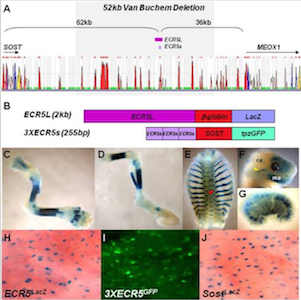
Sost and its paralog Sostdc1 coordinate digit number in a Gli3-dependent manner. Collette NM, Yee CS, Murugesh D, Sebastian A, Taher L, Gale NW, Economides AN, Harland RM, Loots GG. Dev Biol. 2013 Nov 1;383(1):90-105. doi: 10.1016/j.ydbio.2013.08.015. Epub 2013 Aug 29.
The use of nanolipoprotein particles to enhance the immunostimulatory properties of innate immune agonists against lethal influenza challenge. Weilhammer DR, Blanchette CD, Fischer NO, Alam S, Loots GG, Corzett M, Thomas C, Lychak C, Dunkle AD, Ruitenberg JJ, Ghanekar SA, Sant AJ, Rasley A. Biomaterials. 2013;34(38):10305-18. doi: 10.1016/j.biomaterials.2013.09.038.
The use of nanolipoprotein particles to enhance the immunostimulatory properties of innate immune agonists against lethal influenza challenge. Weilhammer DR, Blanchette CD, Fischer NO, Alam S, Loots GG, Corzett M, Thomas C, Lychak C, Dunkle AD, Ruitenberg JJ, Ghanekar SA, Sant AJ, Rasley A. Biomaterials. 2013 Dec;34(38):10305-18. doi: 10.1016/j.biomaterials.2013.09.038. Epub 2013 Sep 27.

Copyright © 2022, Lawrence Livermore National Laboratory